Note
Click here to download the full example code
OT mapping estimation for domain adaptation¶
This example presents how to use MappingTransport to estimate at the same time both the coupling transport and approximate the transport map with either a linear or a kernelized mapping as introduced in [8].
- [8] M. Perrot, N. Courty, R. Flamary, A. Habrard,
“Mapping estimation for discrete optimal transport”, Neural Information Processing Systems (NIPS), 2016.
# Authors: Remi Flamary <remi.flamary@unice.fr>
# Stanislas Chambon <stan.chambon@gmail.com>
#
# License: MIT License
import numpy as np
import matplotlib.pylab as pl
import ot
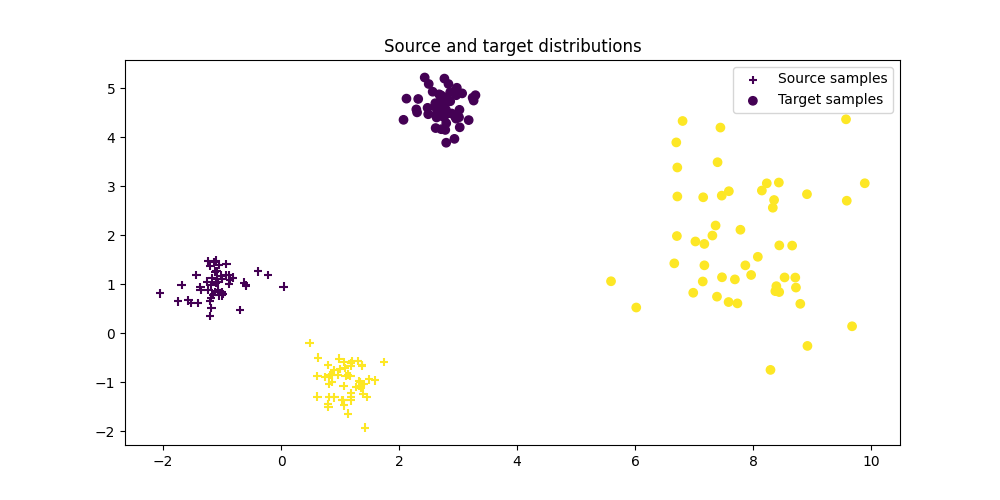
Generate data¶
n_source_samples = 100
n_target_samples = 100
theta = 2 * np.pi / 20
noise_level = 0.1
Xs, ys = ot.datasets.make_data_classif(
'gaussrot', n_source_samples, nz=noise_level)
Xs_new, _ = ot.datasets.make_data_classif(
'gaussrot', n_source_samples, nz=noise_level)
Xt, yt = ot.datasets.make_data_classif(
'gaussrot', n_target_samples, theta=theta, nz=noise_level)
# one of the target mode changes its variance (no linear mapping)
Xt[yt == 2] *= 3
Xt = Xt + 4
Instantiate the different transport algorithms and fit them¶
# MappingTransport with linear kernel
ot_mapping_linear = ot.da.MappingTransport(
kernel="linear", mu=1e0, eta=1e-8, bias=True,
max_iter=20, verbose=True)
ot_mapping_linear.fit(Xs=Xs, Xt=Xt)
# for original source samples, transform applies barycentric mapping
transp_Xs_linear = ot_mapping_linear.transform(Xs=Xs)
# for out of source samples, transform applies the linear mapping
transp_Xs_linear_new = ot_mapping_linear.transform(Xs=Xs_new)
# MappingTransport with gaussian kernel
ot_mapping_gaussian = ot.da.MappingTransport(
kernel="gaussian", eta=1e-5, mu=1e-1, bias=True, sigma=1,
max_iter=10, verbose=True)
ot_mapping_gaussian.fit(Xs=Xs, Xt=Xt)
# for original source samples, transform applies barycentric mapping
transp_Xs_gaussian = ot_mapping_gaussian.transform(Xs=Xs)
# for out of source samples, transform applies the gaussian mapping
transp_Xs_gaussian_new = ot_mapping_gaussian.transform(Xs=Xs_new)
Out:
It. |Loss |Delta loss
--------------------------------
0|4.427565e+03|0.000000e+00
1|4.421944e+03|-1.269514e-03
2|4.421558e+03|-8.726923e-05
3|4.421416e+03|-3.218072e-05
4|4.421340e+03|-1.729876e-05
5|4.421311e+03|-6.394759e-06
It. |Loss |Delta loss
--------------------------------
0|4.431848e+02|0.000000e+00
1|4.412298e+02|-4.411399e-03
2|4.410982e+02|-2.982630e-04
3|4.410234e+02|-1.694337e-04
4|4.409740e+02|-1.121812e-04
5|4.409400e+02|-7.695687e-05
6|4.409119e+02|-6.385031e-05
7|4.408915e+02|-4.611805e-05
8|4.408752e+02|-3.703694e-05
9|4.408620e+02|-2.991123e-05
10|4.408507e+02|-2.559204e-05
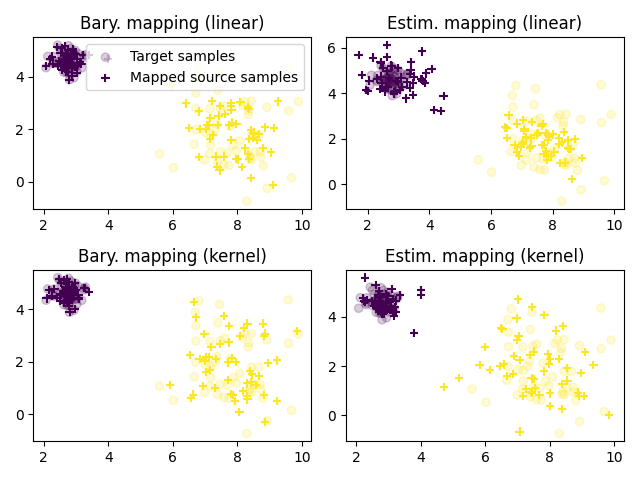
Plot transported samples¶
pl.figure(2)
pl.clf()
pl.subplot(2, 2, 1)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker='o',
label='Target samples', alpha=.2)
pl.scatter(transp_Xs_linear[:, 0], transp_Xs_linear[:, 1], c=ys, marker='+',
label='Mapped source samples')
pl.title("Bary. mapping (linear)")
pl.legend(loc=0)
pl.subplot(2, 2, 2)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker='o',
label='Target samples', alpha=.2)
pl.scatter(transp_Xs_linear_new[:, 0], transp_Xs_linear_new[:, 1],
c=ys, marker='+', label='Learned mapping')
pl.title("Estim. mapping (linear)")
pl.subplot(2, 2, 3)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker='o',
label='Target samples', alpha=.2)
pl.scatter(transp_Xs_gaussian[:, 0], transp_Xs_gaussian[:, 1], c=ys,
marker='+', label='barycentric mapping')
pl.title("Bary. mapping (kernel)")
pl.subplot(2, 2, 4)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker='o',
label='Target samples', alpha=.2)
pl.scatter(transp_Xs_gaussian_new[:, 0], transp_Xs_gaussian_new[:, 1], c=ys,
marker='+', label='Learned mapping')
pl.title("Estim. mapping (kernel)")
pl.tight_layout()
pl.show()
Out:
/home/circleci/project/examples/plot_otda_mapping.py:125: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
pl.show()
Total running time of the script: ( 0 minutes 0.589 seconds)