Note
Go to the end to download the full example code.
Wasserstein Discriminant Analysis
Note
Example added in release: 0.3.0.
This example illustrate the use of WDA as proposed in [11].
[11] Flamary, R., Cuturi, M., Courty, N., & Rakotomamonjy, A. (2016). Wasserstein Discriminant Analysis.
Generate data
n = 1000 # nb samples in source and target datasets
nz = 0.2
np.random.seed(1)
# generate circle dataset
t = np.random.rand(n) * 2 * np.pi
ys = np.floor((np.arange(n) * 1.0 / n * 3)) + 1
xs = np.concatenate((np.cos(t).reshape((-1, 1)), np.sin(t).reshape((-1, 1))), 1)
xs = xs * ys.reshape(-1, 1) + nz * np.random.randn(n, 2)
t = np.random.rand(n) * 2 * np.pi
yt = np.floor((np.arange(n) * 1.0 / n * 3)) + 1
xt = np.concatenate((np.cos(t).reshape((-1, 1)), np.sin(t).reshape((-1, 1))), 1)
xt = xt * yt.reshape(-1, 1) + nz * np.random.randn(n, 2)
nbnoise = 8
xs = np.hstack((xs, np.random.randn(n, nbnoise)))
xt = np.hstack((xt, np.random.randn(n, nbnoise)))
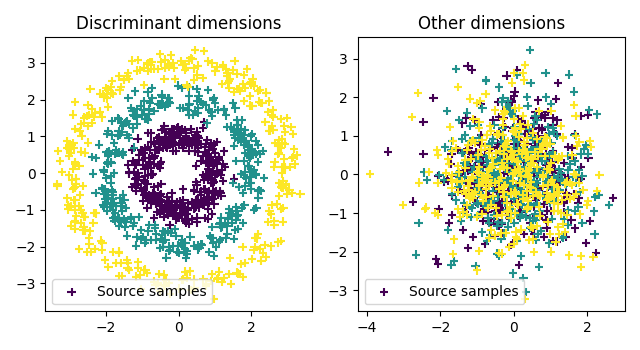
Plot data
pl.figure(1, figsize=(6.4, 3.5))
pl.subplot(1, 2, 1)
pl.scatter(xt[:, 0], xt[:, 1], c=ys, marker="+", label="Source samples")
pl.legend(loc=0)
pl.title("Discriminant dimensions")
pl.subplot(1, 2, 2)
pl.scatter(xt[:, 2], xt[:, 3], c=ys, marker="+", label="Source samples")
pl.legend(loc=0)
pl.title("Other dimensions")
pl.tight_layout()
Compute Fisher Discriminant Analysis
Compute Wasserstein Discriminant Analysis
Optimizing...
Iteration Cost Gradient norm
--------- ----------------------- --------------
1 +8.3042776946697494e-01 5.65147154e-01
2 +4.4401037686381051e-01 2.16760501e-01
3 +4.2234351238819923e-01 1.30555049e-01
4 +4.2169879996364512e-01 1.39115407e-01
5 +4.1924746118060852e-01 1.25387848e-01
6 +4.1177409528991366e-01 6.70993539e-02
7 +4.0862213476138876e-01 3.52716830e-02
8 +4.0747229322240486e-01 3.34923131e-02
9 +4.0678766065260413e-01 2.74029183e-02
10 +4.0621337155460657e-01 2.03651803e-02
11 +4.0577080390746961e-01 2.59605592e-02
12 +4.0543140912490133e-01 3.28883715e-02
13 +4.0470236926315406e-01 1.47528039e-02
14 +4.0445628466113015e-01 5.03183251e-02
15 +4.0364189450997889e-01 3.31006491e-02
16 +4.0303977567984017e-01 1.39885389e-02
17 +4.0301476218564236e-01 2.17467500e-02
18 +4.0292344208896491e-01 1.79959416e-02
19 +4.0271888262078476e-01 6.94410083e-03
20 +4.0183218329658610e-01 1.98336127e-02
21 +3.9762891544683304e-01 1.03191560e-01
22 +3.8226926897284630e-01 1.35962578e-01
23 +3.0859243846661483e-01 1.92704550e-01
24 +2.7991859633835015e-01 2.01770568e-01
25 +2.3708342026018231e-01 9.15797713e-02
26 +2.3380401875457987e-01 6.73647620e-02
27 +2.3061708734620151e-01 4.19289693e-03
28 +2.3061669948481955e-01 4.19499225e-03
29 +2.3061519732125807e-01 3.92852235e-03
30 +2.3061003105938882e-01 2.82794938e-03
31 +2.3060852373964541e-01 2.44254776e-03
32 +2.3060471854906608e-01 6.45973891e-04
33 +2.3060454740611516e-01 4.19467692e-04
34 +2.3060444900856542e-01 1.79947889e-04
35 +2.3060442741354079e-01 2.23617788e-05
36 +2.3060442741310502e-01 2.23518788e-05
37 +2.3060442741136428e-01 2.22940946e-05
38 +2.3060442740443740e-01 2.20626649e-05
39 +2.3060442737731426e-01 2.11320882e-05
40 +2.3060442727858727e-01 1.73278562e-05
41 +2.3060442707611048e-01 4.56119589e-07
Terminated - min grad norm reached after 41 iterations, 7.07 seconds.
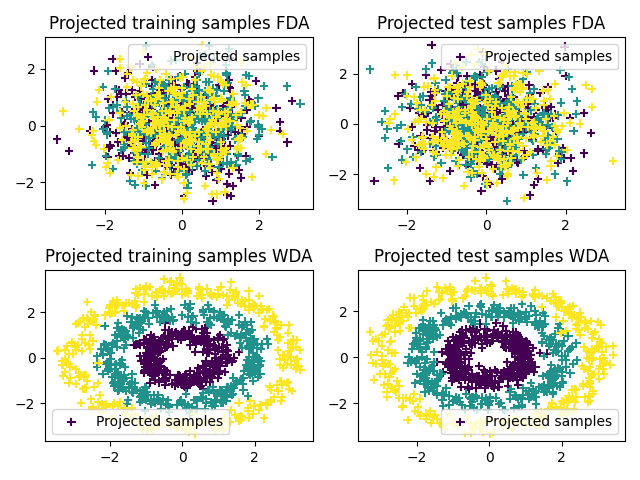
Plot 2D projections
xsp = projfda(xs)
xtp = projfda(xt)
xspw = projwda(xs)
xtpw = projwda(xt)
pl.figure(2)
pl.subplot(2, 2, 1)
pl.scatter(xsp[:, 0], xsp[:, 1], c=ys, marker="+", label="Projected samples")
pl.legend(loc=0)
pl.title("Projected training samples FDA")
pl.subplot(2, 2, 2)
pl.scatter(xtp[:, 0], xtp[:, 1], c=ys, marker="+", label="Projected samples")
pl.legend(loc=0)
pl.title("Projected test samples FDA")
pl.subplot(2, 2, 3)
pl.scatter(xspw[:, 0], xspw[:, 1], c=ys, marker="+", label="Projected samples")
pl.legend(loc=0)
pl.title("Projected training samples WDA")
pl.subplot(2, 2, 4)
pl.scatter(xtpw[:, 0], xtpw[:, 1], c=ys, marker="+", label="Projected samples")
pl.legend(loc=0)
pl.title("Projected test samples WDA")
pl.tight_layout()
pl.show()
/home/circleci/.local/lib/python3.10/site-packages/matplotlib/cbook.py:1719: ComplexWarning: Casting complex values to real discards the imaginary part
return math.isfinite(val)
/home/circleci/.local/lib/python3.10/site-packages/matplotlib/collections.py:200: ComplexWarning: Casting complex values to real discards the imaginary part
offsets = np.asanyarray(offsets, float)
Total running time of the script: (0 minutes 7.810 seconds)