Note
Go to the end to download the full example code.
Gromov-Wasserstein Barycenter example
This example is designed to show how to use the Gromov-Wasserstein distance computation in POT.
# Author: Erwan Vautier <erwan.vautier@gmail.com>
# Nicolas Courty <ncourty@irisa.fr>
#
# License: MIT License
import os
from pathlib import Path
import numpy as np
import scipy as sp
from matplotlib import pyplot as plt
from sklearn import manifold
from sklearn.decomposition import PCA
import ot
Smacof MDS
This function allows to find an embedding of points given a dissimilarity matrix that will be given by the output of the algorithm
def smacof_mds(C, dim, max_iter=3000, eps=1e-9):
"""
Returns an interpolated point cloud following the dissimilarity matrix C
using SMACOF multidimensional scaling (MDS) in specific dimensioned
target space
Parameters
----------
C : ndarray, shape (ns, ns)
dissimilarity matrix
dim : int
dimension of the targeted space
max_iter : int
Maximum number of iterations of the SMACOF algorithm for a single run
eps : float
relative tolerance w.r.t stress to declare converge
Returns
-------
npos : ndarray, shape (R, dim)
Embedded coordinates of the interpolated point cloud (defined with
one isometry)
"""
rng = np.random.RandomState(seed=3)
mds = manifold.MDS(
dim, max_iter=max_iter, eps=1e-9, dissimilarity="precomputed", n_init=1
)
pos = mds.fit(C).embedding_
nmds = manifold.MDS(
2,
max_iter=max_iter,
eps=1e-9,
dissimilarity="precomputed",
random_state=rng,
n_init=1,
)
npos = nmds.fit_transform(C, init=pos)
return npos
Data preparation
The four distributions are constructed from 4 simple images
def im2mat(img):
"""Converts and image to matrix (one pixel per line)"""
return img.reshape((img.shape[0] * img.shape[1], img.shape[2]))
this_file = os.path.realpath("__file__")
data_path = os.path.join(Path(this_file).parent.parent.parent, "data")
square = plt.imread(os.path.join(data_path, "square.png")).astype(np.float64)[:, :, 2]
cross = plt.imread(os.path.join(data_path, "cross.png")).astype(np.float64)[:, :, 2]
triangle = plt.imread(os.path.join(data_path, "triangle.png")).astype(np.float64)[
:, :, 2
]
star = plt.imread(os.path.join(data_path, "star.png")).astype(np.float64)[:, :, 2]
shapes = [square, cross, triangle, star]
S = 4
xs = [[] for i in range(S)]
for nb in range(4):
for i in range(8):
for j in range(8):
if shapes[nb][i, j] < 0.95:
xs[nb].append([j, 8 - i])
xs = [np.array(xs[s]) for s in range(S)]
Barycenter computation
ns = [len(xs[s]) for s in range(S)]
n_samples = 30
"""Compute all distances matrices for the four shapes"""
Cs = [sp.spatial.distance.cdist(xs[s], xs[s]) for s in range(S)]
Cs = [cs / cs.max() for cs in Cs]
ps = [ot.unif(ns[s]) for s in range(S)]
p = ot.unif(n_samples)
lambdast = [[float(i) / 3, float(3 - i) / 3] for i in [1, 2]]
Ct01 = [0 for i in range(2)]
for i in range(2):
Ct01[i] = ot.gromov.gromov_barycenters(
n_samples,
[Cs[0], Cs[1]],
[ps[0], ps[1]],
p,
lambdast[i],
"square_loss", # 5e-4,
max_iter=100,
tol=1e-3,
)
Ct02 = [0 for i in range(2)]
for i in range(2):
Ct02[i] = ot.gromov.gromov_barycenters(
n_samples,
[Cs[0], Cs[2]],
[ps[0], ps[2]],
p,
lambdast[i],
"square_loss", # 5e-4,
max_iter=100,
tol=1e-3,
)
Ct13 = [0 for i in range(2)]
for i in range(2):
Ct13[i] = ot.gromov.gromov_barycenters(
n_samples,
[Cs[1], Cs[3]],
[ps[1], ps[3]],
p,
lambdast[i],
"square_loss", # 5e-4,
max_iter=100,
tol=1e-3,
)
Ct23 = [0 for i in range(2)]
for i in range(2):
Ct23[i] = ot.gromov.gromov_barycenters(
n_samples,
[Cs[2], Cs[3]],
[ps[2], ps[3]],
p,
lambdast[i],
"square_loss", # 5e-4,
max_iter=100,
tol=1e-3,
)
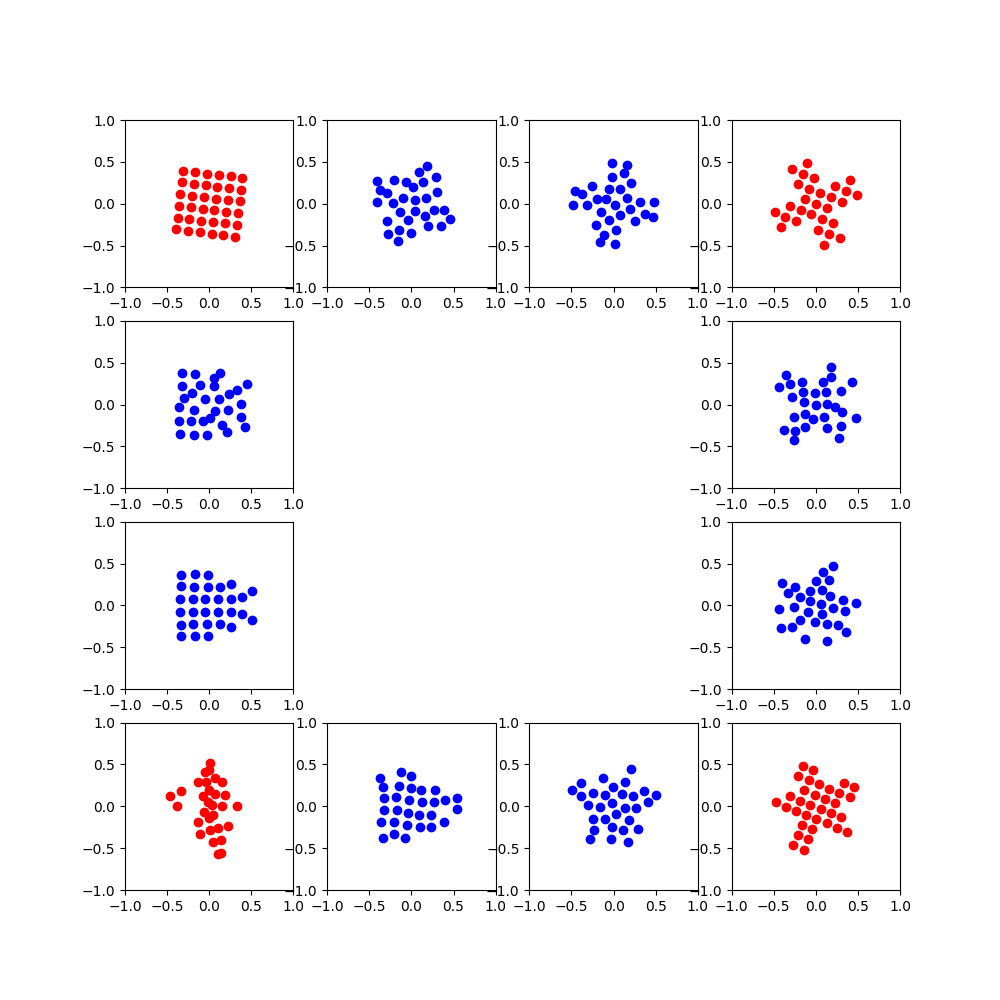
Visualization
The PCA helps in getting consistency between the rotations
clf = PCA(n_components=2)
npos = [0, 0, 0, 0]
npos = [smacof_mds(Cs[s], 2) for s in range(S)]
npost01 = [0, 0]
npost01 = [smacof_mds(Ct01[s], 2) for s in range(2)]
npost01 = [clf.fit_transform(npost01[s]) for s in range(2)]
npost02 = [0, 0]
npost02 = [smacof_mds(Ct02[s], 2) for s in range(2)]
npost02 = [clf.fit_transform(npost02[s]) for s in range(2)]
npost13 = [0, 0]
npost13 = [smacof_mds(Ct13[s], 2) for s in range(2)]
npost13 = [clf.fit_transform(npost13[s]) for s in range(2)]
npost23 = [0, 0]
npost23 = [smacof_mds(Ct23[s], 2) for s in range(2)]
npost23 = [clf.fit_transform(npost23[s]) for s in range(2)]
fig = plt.figure(figsize=(10, 10))
ax1 = plt.subplot2grid((4, 4), (0, 0))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax1.scatter(npos[0][:, 0], npos[0][:, 1], color="r")
ax2 = plt.subplot2grid((4, 4), (0, 1))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax2.scatter(npost01[1][:, 0], npost01[1][:, 1], color="b")
ax3 = plt.subplot2grid((4, 4), (0, 2))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax3.scatter(npost01[0][:, 0], npost01[0][:, 1], color="b")
ax4 = plt.subplot2grid((4, 4), (0, 3))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax4.scatter(npos[1][:, 0], npos[1][:, 1], color="r")
ax5 = plt.subplot2grid((4, 4), (1, 0))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax5.scatter(npost02[1][:, 0], npost02[1][:, 1], color="b")
ax6 = plt.subplot2grid((4, 4), (1, 3))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax6.scatter(npost13[1][:, 0], npost13[1][:, 1], color="b")
ax7 = plt.subplot2grid((4, 4), (2, 0))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax7.scatter(npost02[0][:, 0], npost02[0][:, 1], color="b")
ax8 = plt.subplot2grid((4, 4), (2, 3))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax8.scatter(npost13[0][:, 0], npost13[0][:, 1], color="b")
ax9 = plt.subplot2grid((4, 4), (3, 0))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax9.scatter(npos[2][:, 0], npos[2][:, 1], color="r")
ax10 = plt.subplot2grid((4, 4), (3, 1))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax10.scatter(npost23[1][:, 0], npost23[1][:, 1], color="b")
ax11 = plt.subplot2grid((4, 4), (3, 2))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax11.scatter(npost23[0][:, 0], npost23[0][:, 1], color="b")
ax12 = plt.subplot2grid((4, 4), (3, 3))
plt.xlim((-1, 1))
plt.ylim((-1, 1))
ax12.scatter(npos[3][:, 0], npos[3][:, 1], color="r")
<matplotlib.collections.PathCollection object at 0x7f95b593c280>
Total running time of the script: (0 minutes 1.945 seconds)