Note
Go to the end to download the full example code.
Barycenter of labeled graphs with FGW
This example illustrates the computation barycenter of labeled graphs using FGW [18].
Requires networkx >=2
[18] Vayer Titouan, Chapel Laetitia, Flamary Rémi, Tavenard Romain and Courty Nicolas “Optimal Transport for structured data with application on graphs” International Conference on Machine Learning (ICML). 2019.
# Author: Titouan Vayer <titouan.vayer@irisa.fr>
#
# License: MIT License
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
import math
from scipy.sparse.csgraph import shortest_path
import matplotlib.colors as mcol
from matplotlib import cm
from ot.gromov import fgw_barycenters
def find_thresh(C, inf=0.5, sup=3, step=10):
"""Trick to find the adequate thresholds from where value of the C matrix are considered close enough to say that nodes are connected
The threshold is found by a linesearch between values "inf" and "sup" with "step" thresholds tested.
The optimal threshold is the one which minimizes the reconstruction error between the shortest_path matrix coming from the thresholded adjacency matrix
and the original matrix.
Parameters
----------
C : ndarray, shape (n_nodes,n_nodes)
The structure matrix to threshold
inf : float
The beginning of the linesearch
sup : float
The end of the linesearch
step : integer
Number of thresholds tested
"""
dist = []
search = np.linspace(inf, sup, step)
for thresh in search:
Cprime = sp_to_adjacency(C, 0, thresh)
SC = shortest_path(Cprime, method="D")
SC[SC == float("inf")] = 100
dist.append(np.linalg.norm(SC - C))
return search[np.argmin(dist)], dist
def sp_to_adjacency(C, threshinf=0.2, threshsup=1.8):
"""Thresholds the structure matrix in order to compute an adjacency matrix.
All values between threshinf and threshsup are considered representing connected nodes and set to 1. Else are set to 0
Parameters
----------
C : ndarray, shape (n_nodes,n_nodes)
The structure matrix to threshold
threshinf : float
The minimum value of distance from which the new value is set to 1
threshsup : float
The maximum value of distance from which the new value is set to 1
Returns
-------
C : ndarray, shape (n_nodes,n_nodes)
The threshold matrix. Each element is in {0,1}
"""
H = np.zeros_like(C)
np.fill_diagonal(H, np.diagonal(C))
C = C - H
C = np.minimum(np.maximum(C, threshinf), threshsup)
C[C == threshsup] = 0
C[C != 0] = 1
return C
def build_noisy_circular_graph(
N=20, mu=0, sigma=0.3, with_noise=False, structure_noise=False, p=None
):
"""Create a noisy circular graph"""
g = nx.Graph()
g.add_nodes_from(list(range(N)))
for i in range(N):
noise = np.random.normal(mu, sigma, 1)[0]
if with_noise:
g.add_node(i, attr_name=math.sin((2 * i * math.pi / N)) + noise)
else:
g.add_node(i, attr_name=math.sin(2 * i * math.pi / N))
g.add_edge(i, i + 1)
if structure_noise:
randomint = np.random.randint(0, p)
if randomint == 0:
if i <= N - 3:
g.add_edge(i, i + 2)
if i == N - 2:
g.add_edge(i, 0)
if i == N - 1:
g.add_edge(i, 1)
g.add_edge(N, 0)
noise = np.random.normal(mu, sigma, 1)[0]
if with_noise:
g.add_node(N, attr_name=math.sin((2 * N * math.pi / N)) + noise)
else:
g.add_node(N, attr_name=math.sin(2 * N * math.pi / N))
return g
def graph_colors(nx_graph, vmin=0, vmax=7):
cnorm = mcol.Normalize(vmin=vmin, vmax=vmax)
cpick = cm.ScalarMappable(norm=cnorm, cmap="viridis")
cpick.set_array([])
val_map = {}
for k, v in nx.get_node_attributes(nx_graph, "attr_name").items():
val_map[k] = cpick.to_rgba(v)
colors = []
for node in nx_graph.nodes():
colors.append(val_map[node])
return colors
Generate data
We build a dataset of noisy circular graphs. Noise is added on the structures by random connections and on the features by gaussian noise.
np.random.seed(30)
X0 = []
for k in range(9):
X0.append(
build_noisy_circular_graph(
np.random.randint(15, 25), with_noise=True, structure_noise=True, p=3
)
)
Plot data
plt.figure(figsize=(8, 10))
for i in range(len(X0)):
plt.subplot(3, 3, i + 1)
g = X0[i]
pos = nx.kamada_kawai_layout(g)
nx.draw_networkx(
g,
pos=pos,
node_color=graph_colors(g, vmin=-1, vmax=1),
with_labels=False,
node_size=100,
)
plt.suptitle("Dataset of noisy graphs. Color indicates the label", fontsize=20)
plt.show()
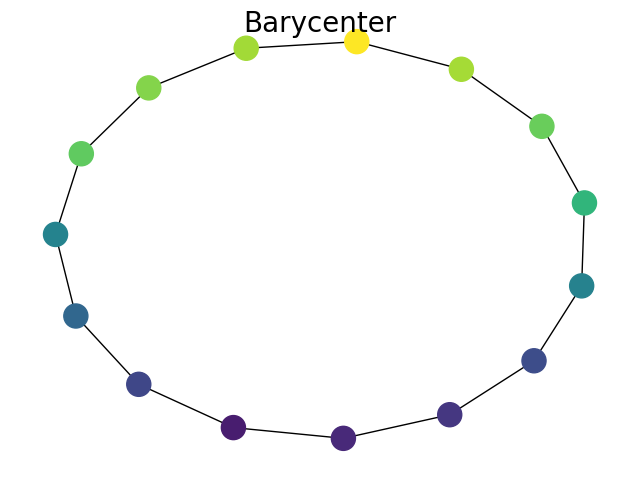
Barycenter computation
Features distances are the euclidean distances
Cs = [shortest_path(nx.adjacency_matrix(x).toarray()) for x in X0]
ps = [np.ones(len(x.nodes())) / len(x.nodes()) for x in X0]
Ys = [
np.array([v for (k, v) in nx.get_node_attributes(x, "attr_name").items()]).reshape(
-1, 1
)
for x in X0
]
lambdas = np.array([np.ones(len(Ys)) / len(Ys)]).ravel()
sizebary = 15 # we choose a barycenter with 15 nodes
A, C, log = fgw_barycenters(sizebary, Ys, Cs, ps, lambdas, alpha=0.95, log=True)
Plot Barycenter
pos = nx.kamada_kawai_layout(bary)
nx.draw_networkx(
bary, pos=pos, node_color=graph_colors(bary, vmin=-1, vmax=1), with_labels=False
)
plt.suptitle("Barycenter", fontsize=20)
plt.show()
Total running time of the script: (0 minutes 1.615 seconds)