Note
Go to the end to download the full example code.
1D Wasserstein barycenter: exact LP vs entropic regularization
This example illustrates the computation of regularized Wasserstein Barycenter as proposed in [3] and exact LP barycenters using standard LP solver.
It reproduces approximately Figure 3.1 and 3.2 from the following paper: Cuturi, M., & Peyré, G. (2016). A smoothed dual approach for variational Wasserstein problems. SIAM Journal on Imaging Sciences, 9(1), 320-343.
[3] Benamou, J. D., Carlier, G., Cuturi, M., Nenna, L., & Peyré, G. (2015). Iterative Bregman projections for regularized transportation problems SIAM Journal on Scientific Computing, 37(2), A1111-A1138.
# Author: Remi Flamary <remi.flamary@unice.fr>
#
# License: MIT License
# sphinx_gallery_thumbnail_number = 4
import numpy as np
import matplotlib.pylab as pl
import ot
# necessary for 3d plot even if not used
from mpl_toolkits.mplot3d import Axes3D # noqa
from matplotlib.collections import PolyCollection # noqa
# import ot.lp.cvx as cvx
Gaussian Data
problems = []
n = 100 # nb bins
# bin positions
x = np.arange(n, dtype=np.float64)
# Gaussian distributions
# Gaussian distributions
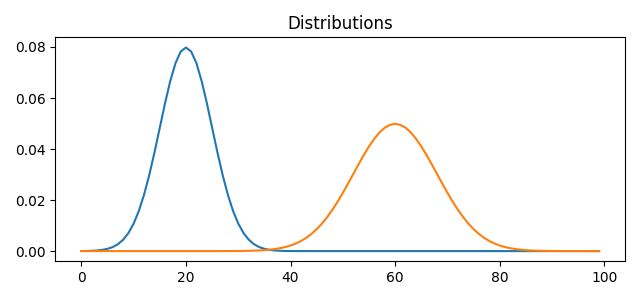
a1 = ot.datasets.make_1D_gauss(n, m=20, s=5) # m= mean, s= std
a2 = ot.datasets.make_1D_gauss(n, m=60, s=8)
# creating matrix A containing all distributions
A = np.vstack((a1, a2)).T
n_distributions = A.shape[1]
# loss matrix + normalization
M = ot.utils.dist0(n)
M /= M.max()
pl.figure(1, figsize=(6.4, 3))
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.tight_layout()
alpha = 0.5 # 0<=alpha<=1
weights = np.array([1 - alpha, alpha])
# l2bary
bary_l2 = A.dot(weights)
# wasserstein
reg = 1e-3
ot.tic()
bary_wass = ot.bregman.barycenter(A, M, reg, weights)
ot.toc()
ot.tic()
bary_wass2 = ot.lp.barycenter(A, M, weights)
ot.toc()
pl.figure(2)
pl.clf()
pl.subplot(2, 1, 1)
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.subplot(2, 1, 2)
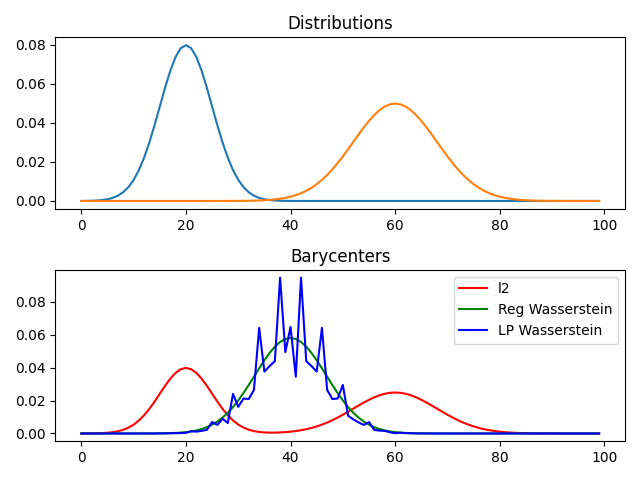
pl.plot(x, bary_l2, "r", label="l2")
pl.plot(x, bary_wass, "g", label="Reg Wasserstein")
pl.plot(x, bary_wass2, "b", label="LP Wasserstein")
pl.legend()
pl.title("Barycenters")
pl.tight_layout()
problems.append([A, [bary_l2, bary_wass, bary_wass2]])
Elapsed time : 0.0035399970001890324 s
Elapsed time : 0.26590077599985307 s
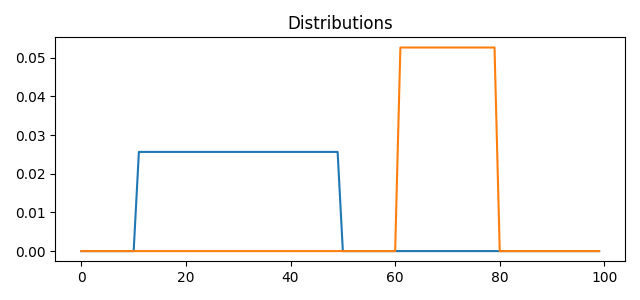
Stair Data
pl.figure(1, figsize=(6.4, 3))
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.tight_layout()
alpha = 0.5 # 0<=alpha<=1
weights = np.array([1 - alpha, alpha])
# l2bary
bary_l2 = A.dot(weights)
# wasserstein
reg = 1e-3
ot.tic()
bary_wass = ot.bregman.barycenter(A, M, reg, weights)
ot.toc()
ot.tic()
bary_wass2 = ot.lp.barycenter(A, M, weights)
ot.toc()
problems.append([A, [bary_l2, bary_wass, bary_wass2]])
pl.figure(2)
pl.clf()
pl.subplot(2, 1, 1)
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.subplot(2, 1, 2)
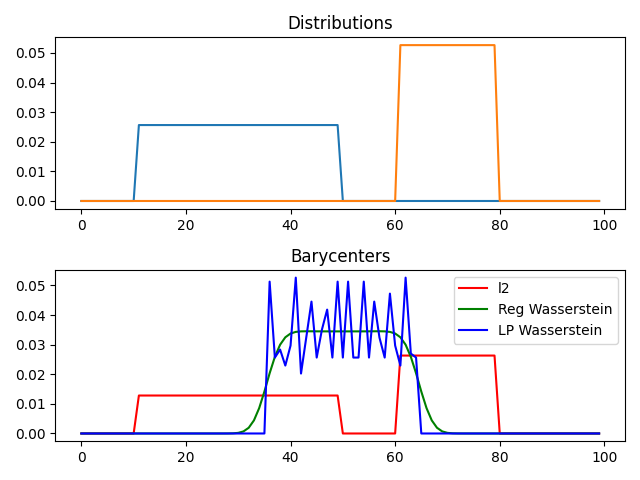
pl.plot(x, bary_l2, "r", label="l2")
pl.plot(x, bary_wass, "g", label="Reg Wasserstein")
pl.plot(x, bary_wass2, "b", label="LP Wasserstein")
pl.legend()
pl.title("Barycenters")
pl.tight_layout()
Elapsed time : 0.005050758000379574 s
Elapsed time : 0.10086229799981083 s
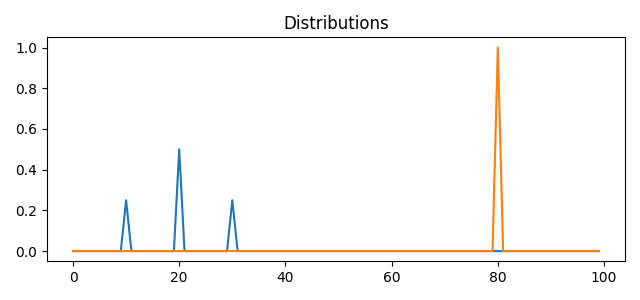
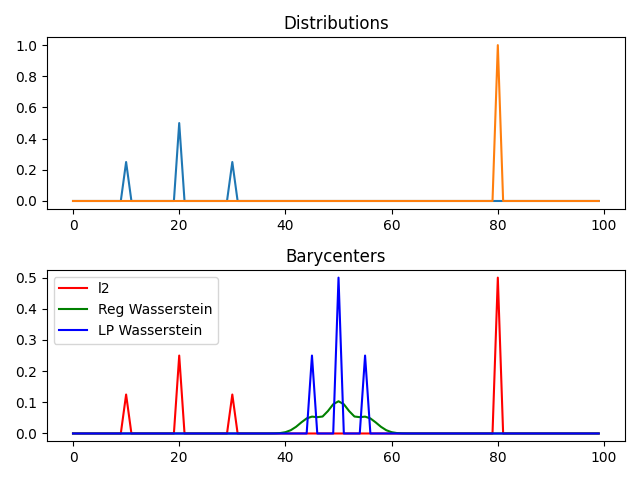
Dirac Data
pl.figure(1, figsize=(6.4, 3))
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.tight_layout()
alpha = 0.5 # 0<=alpha<=1
weights = np.array([1 - alpha, alpha])
# l2bary
bary_l2 = A.dot(weights)
# wasserstein
reg = 1e-3
ot.tic()
bary_wass = ot.bregman.barycenter(A, M, reg, weights)
ot.toc()
ot.tic()
bary_wass2 = ot.lp.barycenter(A, M, weights)
ot.toc()
problems.append([A, [bary_l2, bary_wass, bary_wass2]])
pl.figure(2)
pl.clf()
pl.subplot(2, 1, 1)
for i in range(n_distributions):
pl.plot(x, A[:, i])
pl.title("Distributions")
pl.subplot(2, 1, 2)
pl.plot(x, bary_l2, "r", label="l2")
pl.plot(x, bary_wass, "g", label="Reg Wasserstein")
pl.plot(x, bary_wass2, "b", label="LP Wasserstein")
pl.legend()
pl.title("Barycenters")
pl.tight_layout()
Elapsed time : 0.0010897010001826857 s
Elapsed time : 0.07999342999983128 s
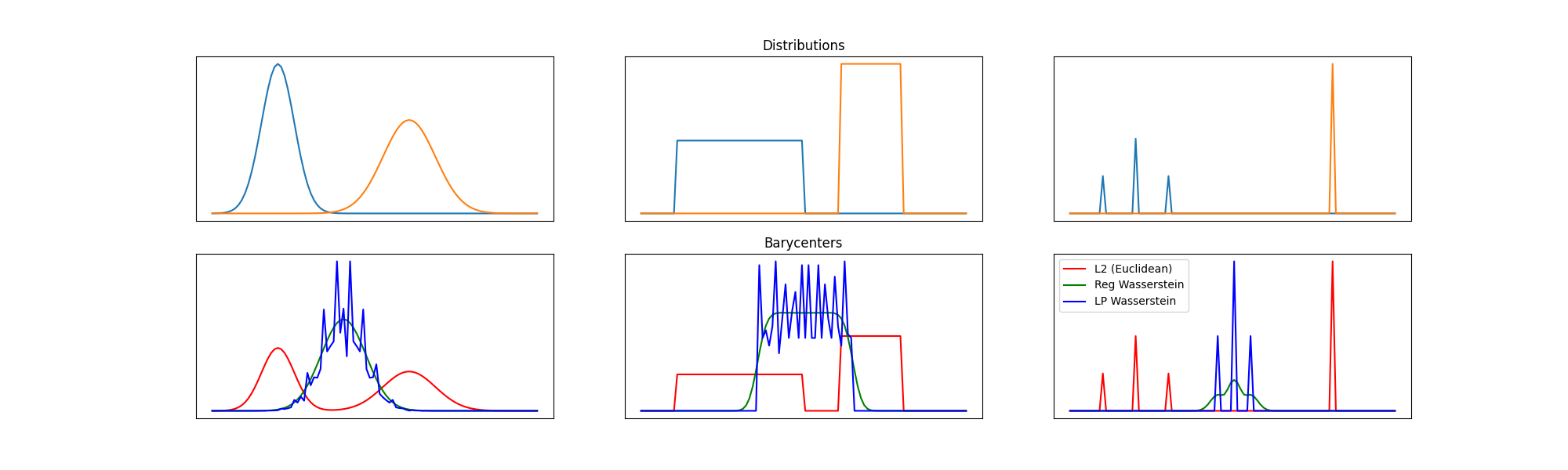
Final figure
nbm = len(problems)
nbm2 = nbm // 2
pl.figure(2, (20, 6))
pl.clf()
for i in range(nbm):
A = problems[i][0]
bary_l2 = problems[i][1][0]
bary_wass = problems[i][1][1]
bary_wass2 = problems[i][1][2]
pl.subplot(2, nbm, 1 + i)
for j in range(n_distributions):
pl.plot(x, A[:, j])
if i == nbm2:
pl.title("Distributions")
pl.xticks(())
pl.yticks(())
pl.subplot(2, nbm, 1 + i + nbm)
pl.plot(x, bary_l2, "r", label="L2 (Euclidean)")
pl.plot(x, bary_wass, "g", label="Reg Wasserstein")
pl.plot(x, bary_wass2, "b", label="LP Wasserstein")
if i == nbm - 1:
pl.legend()
if i == nbm2:
pl.title("Barycenters")
pl.xticks(())
pl.yticks(())
Total running time of the script: (0 minutes 1.695 seconds)