Note
Go to the end to download the full example code.
OT with Laplacian regularization for domain adaptation
Note
Example added in release: 0.7.0.
This example introduces a domain adaptation in a 2D setting and OTDA approach with Laplacian regularization.
# Authors: Ievgen Redko <ievgen.redko@univ-st-etienne.fr>
# License: MIT License
import matplotlib.pylab as pl
import ot
Generate data
n_source_samples = 150
n_target_samples = 150
Xs, ys = ot.datasets.make_data_classif("3gauss", n_source_samples)
Xt, yt = ot.datasets.make_data_classif("3gauss2", n_target_samples)
Instantiate the different transport algorithms and fit them
# EMD Transport
ot_emd = ot.da.EMDTransport()
ot_emd.fit(Xs=Xs, Xt=Xt)
# Sinkhorn Transport
ot_sinkhorn = ot.da.SinkhornTransport(reg_e=0.01)
ot_sinkhorn.fit(Xs=Xs, Xt=Xt)
# EMD Transport with Laplacian regularization
ot_emd_laplace = ot.da.EMDLaplaceTransport(reg_lap=100, reg_src=1)
ot_emd_laplace.fit(Xs=Xs, Xt=Xt)
# transport source samples onto target samples
transp_Xs_emd = ot_emd.transform(Xs=Xs)
transp_Xs_sinkhorn = ot_sinkhorn.transform(Xs=Xs)
transp_Xs_emd_laplace = ot_emd_laplace.transform(Xs=Xs)
/home/circleci/project/ot/bregman/_sinkhorn.py:902: UserWarning: Sinkhorn did not converge. You might want to increase the number of iterations `numItermax` or the regularization parameter `reg`.
warnings.warn(
/home/circleci/project/ot/backend.py:1261: RuntimeWarning: overflow encountered in exp
return np.exp(a)
Fig 1 : plots source and target samples
pl.figure(1, figsize=(10, 5))
pl.subplot(1, 2, 1)
pl.scatter(Xs[:, 0], Xs[:, 1], c=ys, marker="+", label="Source samples")
pl.xticks([])
pl.yticks([])
pl.legend(loc=0)
pl.title("Source samples")
pl.subplot(1, 2, 2)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker="o", label="Target samples")
pl.xticks([])
pl.yticks([])
pl.legend(loc=0)
pl.title("Target samples")
pl.tight_layout()
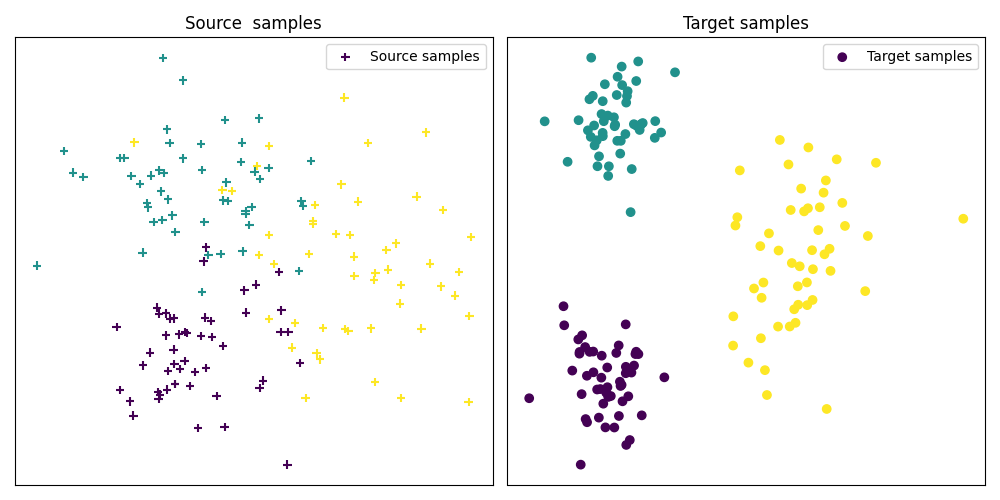
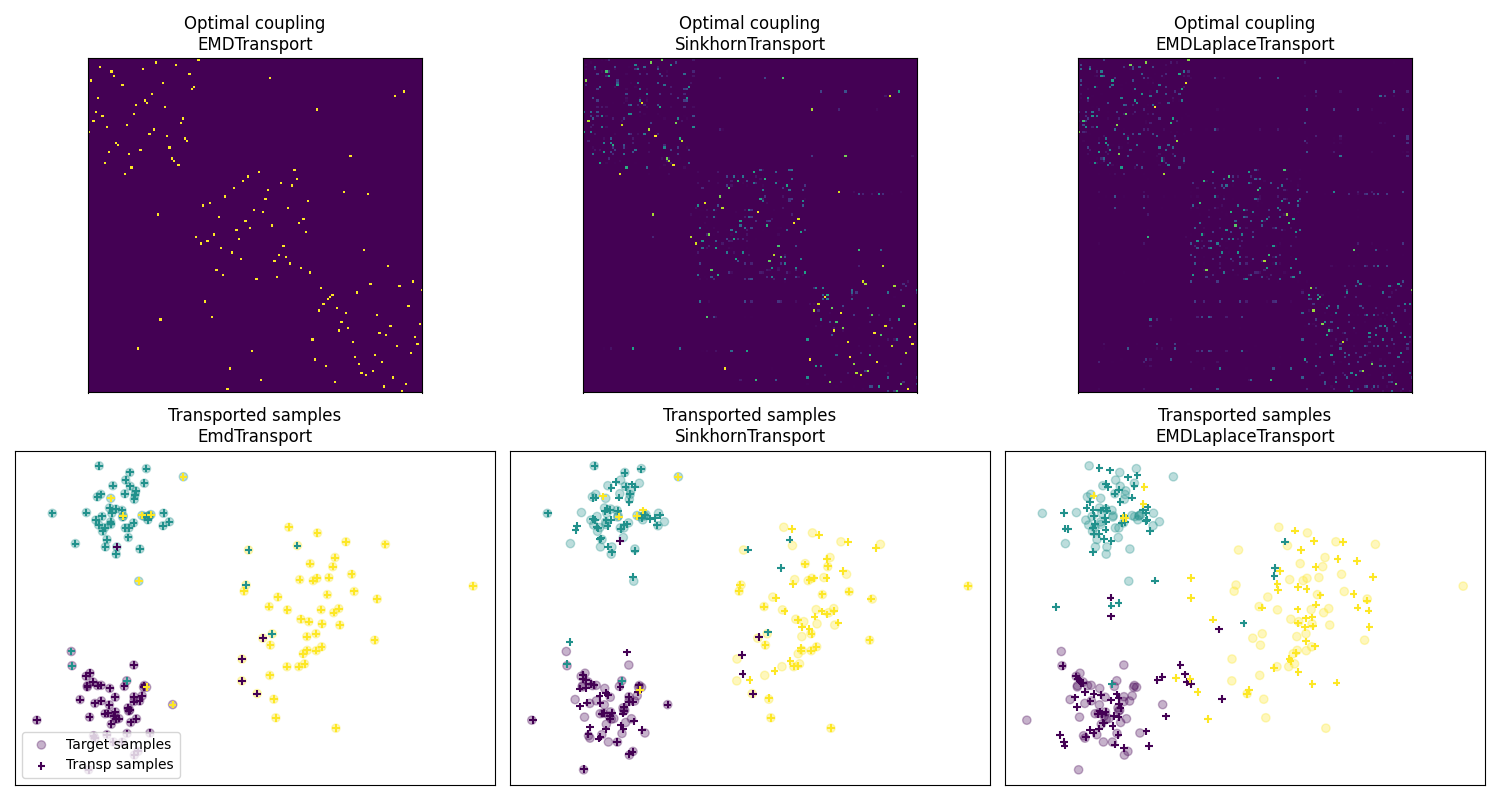
Fig 2 : plot optimal couplings and transported samples
param_img = {"interpolation": "nearest", "cmap": "gray_r"}
pl.figure(2, figsize=(15, 8))
pl.subplot(2, 3, 1)
pl.imshow(ot_emd.coupling_, **param_img)
pl.xticks([])
pl.yticks([])
pl.title("Optimal coupling\nEMDTransport")
pl.figure(2, figsize=(15, 8))
pl.subplot(2, 3, 2)
pl.imshow(ot_sinkhorn.coupling_, **param_img)
pl.xticks([])
pl.yticks([])
pl.title("Optimal coupling\nSinkhornTransport")
pl.subplot(2, 3, 3)
pl.imshow(ot_emd_laplace.coupling_, **param_img)
pl.xticks([])
pl.yticks([])
pl.title("Optimal coupling\nEMDLaplaceTransport")
pl.subplot(2, 3, 4)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker="o", label="Target samples", alpha=0.3)
pl.scatter(
transp_Xs_emd[:, 0],
transp_Xs_emd[:, 1],
c=ys,
marker="+",
label="Transp samples",
s=30,
)
pl.xticks([])
pl.yticks([])
pl.title("Transported samples\nEmdTransport")
pl.legend(loc="lower left")
pl.subplot(2, 3, 5)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker="o", label="Target samples", alpha=0.3)
pl.scatter(
transp_Xs_sinkhorn[:, 0],
transp_Xs_sinkhorn[:, 1],
c=ys,
marker="+",
label="Transp samples",
s=30,
)
pl.xticks([])
pl.yticks([])
pl.title("Transported samples\nSinkhornTransport")
pl.subplot(2, 3, 6)
pl.scatter(Xt[:, 0], Xt[:, 1], c=yt, marker="o", label="Target samples", alpha=0.3)
pl.scatter(
transp_Xs_emd_laplace[:, 0],
transp_Xs_emd_laplace[:, 1],
c=ys,
marker="+",
label="Transp samples",
s=30,
)
pl.xticks([])
pl.yticks([])
pl.title("Transported samples\nEMDLaplaceTransport")
pl.tight_layout()
pl.show()
/home/circleci/project/examples/domain-adaptation/plot_otda_laplacian.py:88: UserWarning: Ignoring specified arguments in this call because figure with num: 2 already exists
pl.figure(2, figsize=(15, 8))
Total running time of the script: (0 minutes 2.428 seconds)